Detector distortion corrections¶
This tutorial shows how to correct images for spatial distortion. Some tutorial examples rely on files available in http://www.silx.org/pub/pyFAI/testimages/ and will be downloaded during this tutorial. The requiered minimum version of pyFAI is 0.12.0 (currently dev5)
Detector definitions¶
PyFAI features an impressive list of 55 detector definitions contributed often by manufacturers and some other reverse engineerd by scientists. Each of them is defined as an invividual class which contains a way to calculate the mask (invalid pixels, gaps,…) and a method to calculate the pixel positions in Cartesian coordinates.
import pyFAI
all_detectors = list(set(pyFAI.detectors.ALL_DETECTORS.values()))
#Sort detectors according to their name
all_detectors.sort(key=lambda i:i.__name__)
nb_det = len(all_detectors)
print("Number of detectors registered:", nb_det)
for i in all_detectors:
print(i())
WARNING:xsdimage:lxml library is probably not part of your python installation: disabling xsdimage format
WARNING:pyFAI.utils:Exception No module named 'fftw3': FFTw3 not available. Falling back on Scipy
Number of detectors registered: 55
Detector Quantum 210 Spline= None PixelSize= 5.100e-05, 5.100e-05 m
Detector Quantum 270 Spline= None PixelSize= 6.480e-05, 6.480e-05 m
Detector Quantum 315 Spline= None PixelSize= 5.100e-05, 5.100e-05 m
Detector Quantum 4 Spline= None PixelSize= 8.200e-05, 8.200e-05 m
Detector Aarhus Spline= None PixelSize= 2.500e-05, 2.500e-05 m
Detector ApexII PixelSize= 6.000e-05, 6.000e-05 m
Detector aca1300 PixelSize= 3.750e-06, 3.750e-06 m
Undefined detector
Detector Dexela 2923 PixelSize= 7.500e-05, 7.500e-05 m
Detector Eiger16M PixelSize= 7.500e-05, 7.500e-05 m
Detector Eiger1M PixelSize= 7.500e-05, 7.500e-05 m
Detector Eiger4M PixelSize= 7.500e-05, 7.500e-05 m
Detector Eiger9M PixelSize= 7.500e-05, 7.500e-05 m
Detector Fairchild PixelSize= 1.500e-05, 1.500e-05 m
Detector HF-130k Spline= None PixelSize= 1.500e-04, 1.500e-04 m
Detector HF-1M Spline= None PixelSize= 1.500e-04, 1.500e-04 m
Detector HF-262k Spline= None PixelSize= 1.500e-04, 1.500e-04 m
Detector HF-2.4M Spline= None PixelSize= 1.500e-04, 1.500e-04 m
Detector HF-4M Spline= None PixelSize= 1.500e-04, 1.500e-04 m
Detector HF-9.4M Spline= None PixelSize= 1.500e-04, 1.500e-04 m
Detector Imxpad S10 PixelSize= 1.300e-04, 1.300e-04 m
Detector Imxpad S140 PixelSize= 1.300e-04, 1.300e-04 m
Detector Imxpad S70 PixelSize= 1.300e-04, 1.300e-04 m
Detector MAR 345 PixelSize= 1.000e-04, 1.000e-04 m
Detector Pixium 4700 detector PixelSize= 1.540e-04, 1.540e-04 m
Detector Perkin detector PixelSize= 2.000e-04, 2.000e-04 m
Detector Pilatus100k PixelSize= 1.720e-04, 1.720e-04 m
Detector Pilatus1M PixelSize= 1.720e-04, 1.720e-04 m
Detector Pilatus200k PixelSize= 1.720e-04, 1.720e-04 m
Detector Pilatus2M PixelSize= 1.720e-04, 1.720e-04 m
Detector Pilatus300k PixelSize= 1.720e-04, 1.720e-04 m
Detector Pilatus300kw PixelSize= 1.720e-04, 1.720e-04 m
Detector Pilatus6M PixelSize= 1.720e-04, 1.720e-04 m
Detector PilatusCdTe1M PixelSize= 1.720e-04, 1.720e-04 m
Detector PilatusCdTe2M PixelSize= 1.720e-04, 1.720e-04 m
Detector PilatusCdTe300k PixelSize= 1.720e-04, 1.720e-04 m
Detector PilatusCdTe300kw PixelSize= 1.720e-04, 1.720e-04 m
Detector Rayonix PixelSize= 3.200e-05, 3.200e-05 m
Detector MAR133 PixelSize= 6.400e-05, 6.400e-05 m
Detector Rayonix lx170 PixelSize= 4.427e-05, 4.427e-05 m
Detector Rayonix lx255 PixelSize= 4.427e-05, 4.427e-05 m
Detector Rayonix mx170 PixelSize= 4.427e-05, 4.427e-05 m
Detector Rayonix mx225 PixelSize= 7.324e-05, 7.324e-05 m
Detector Rayonix mx225hs PixelSize= 7.813e-05, 7.813e-05 m
Detector Rayonix mx300 PixelSize= 7.324e-05, 7.324e-05 m
Detector Rayonix mx300hs PixelSize= 7.813e-05, 7.813e-05 m
Detector Rayonix mx325 PixelSize= 7.935e-05, 7.935e-05 m
Detector Rayonix mx340hs PixelSize= 8.854e-05, 8.854e-05 m
Detector Rayonix mx425hs PixelSize= 4.427e-05, 4.427e-05 m
Detector MAR165 PixelSize= 3.950e-05, 3.950e-05 m
Detector Rayonix sx200 PixelSize= 4.800e-05, 4.800e-05 m
Detector Rayonix Sx30hs PixelSize= 1.563e-05, 1.563e-05 m
Detector Rayonix Sx85hs PixelSize= 4.427e-05, 4.427e-05 m
Detector Titan 2k x 2k PixelSize= 6.000e-05, 6.000e-05 m
Detector Xpad S540 flat PixelSize= 1.300e-04, 1.300e-04 m
Defining a detector from a spline file¶
For optically coupled CCD detectors, the geometrical distortion is often described by a two-dimensional cubic spline (as in FIT2D) which can be imported into the relevant detector instance and used to calculate the actual pixel position in space (and masked pixels).
At the ESRF, mainly FReLoN detectors [J.-C. Labiche, ESRF Newsletter 25, 41 (1996)] are used with spline files describing the distortion of the fiber optic taper.
Let’s download such a file and create a detector from it.
import os, sys
os.environ["http_proxy"] = "http://proxy.site.com:3128"
def download(url):
"""download the file given in URL and return its local path"""
if sys.version_info[0]<3:
from urllib2 import urlopen, ProxyHandler, build_opener
else:
from urllib.request import urlopen, ProxyHandler, build_opener
dictProxies = {}
if "http_proxy" in os.environ:
dictProxies['http'] = os.environ["http_proxy"]
dictProxies['https'] = os.environ["http_proxy"]
if "https_proxy" in os.environ:
dictProxies['https'] = os.environ["https_proxy"]
if dictProxies:
proxy_handler = ProxyHandler(dictProxies)
opener = build_opener(proxy_handler).open
else:
opener = urlopen
target = os.path.split(url)[-1]
with open(target,"wb") as dest, opener(url) as src:
dest.write(src.read())
return target
spline_file = download("http://www.silx.org/pub/pyFAI/testimages/halfccd.spline")
hd = pyFAI.detectors.FReLoN(splineFile=spline_file)
print(hd)
print("Shape: %i, %i"% hd.shape)
Detector FReLoN Spline= /users/kieffer/workspace-400/pyFAI/doc/source/usage/tutorial/Distortion/halfccd.spline PixelSize= 4.842e-05, 4.684e-05 m
Shape: 1025, 2048
Note the unusual shape of this detector. This is probably a human error when calibrating the detector distortion in FIT2D.
Visualizing the mask¶
Every detector object contains a mask attribute, defining pixels which are invalid. For FReLoN detector (a spline-files-defined detectors), all pixels having an offset such that the pixel falls out of the initial detector are considered as invalid.
Masked pixel have non-null values can be displayed like this:
%pylab inline
imshow(hd.mask, origin="lower", interpolation="nearest")
Populating the interactive namespace from numpy and matplotlib
<matplotlib.image.AxesImage at 0x7f2304a8cd68>

Detector definition files as NeXus files¶
Any detector object in pyFAI can be saved into an HDF5 file following the NeXus convention [Könnecke et al., 2015, J. Appl. Cryst. 48, 301-305.]. Detector objects can subsequently be restored from disk, making complex detector definitions less error prone.
h5_file = "halfccd.h5"
hd.save(h5_file)
new_det = pyFAI.detector_factory(h5_file)
print(new_det)
print("Mask is the same: ", numpy.allclose(new_det.mask, hd.mask))
print("Pixel positions are the same: ", numpy.allclose(new_det.get_pixel_corners(), hd.get_pixel_corners()))
print("Number of masked pixels", new_det.mask.sum())
FReLoN detector from NeXus file: halfccd.h5 PixelSize= 4.842e-05, 4.684e-05 m
Mask is the same: True
Pixel positions are the same: True
Number of masked pixels 34382
Pixels of an area detector are saved as a four-dimensional dataset: i.e. a two-dimensional array of vertices pointing to every corner of each pixel, generating an array of dimension (Ny, Nx, Nc, 3), where Nx and Ny are the dimensions of the detector, Nc is the number of corners of each pixel, usually four, and the last entry contains the coordinates of the vertex itself (in the order: Z, Y, X).
This kind of definition, while relying on large description files, can address some of the most complex detector layouts. They will be presented a bit later in this tutorial.
print("Size of Spline-file:", os.stat('halfccd.spline').st_size)
print("Size of Nexus-file:", os.stat('halfccd.h5').st_size)
Size of Spline-file: 1183
Size of Nexus-file: 21451707
The HDF5 file is indeed much larger than the spline file.
Modify a detector and saving¶
One may want to define a new mask (or flat-field) for its detector and save the mask with the detector definition. Here, we create a copy of the detector and reset its mask to enable all pixels in the detector and save the new detector instance into another file.
import copy
nomask_file = "nomask.h5"
nomask = copy.deepcopy(new_det)
nomask.mask = numpy.zeros_like(new_det.mask)
nomask.save(nomask_file)
nomask = pyFAI.detector_factory("nomask.h5")
print("No pixels are masked",nomask.mask.sum())
No pixels are masked 0
Wrap up
In this section we have seen how detectors are defined in pyFAI, how they can be created, either from the list of the parametrized ones, or from spline files, or from NeXus detector files. We have also seen how to save and subsequently restore a detector instance, preserving the modifications made.
Distortion correction¶
Once the position of every single pixel in space is known, one can benefit from the regridding engine of pyFAI adapted to image distortion correction tasks. The pyFAI.distortion.Distortion class is the equivalent of the pyFAI.AzimuthalIntegrator for distortion. Provided with a detector definition, it enables the correction of a set of images by using the same kind of look-up tables as for azimuthal integration.
from pyFAI.distortion import Distortion
dis = Distortion(nomask)
print(dis)
Distortion correction lut on device None for detector shape (1025, 2048):
NexusDetector detector from NeXus file: nomask.h5 PixelSize= 4.842e-05, 4.684e-05 m
FReLoN detector¶
First load the image to be corrected, then correct it for geometric distortion.
halfccd_img = download("http://www.silx.org/pub/pyFAI/testimages/halfccd.edf")
import fabio
raw = fabio.open(halfccd_img).data
cor = dis.correct(raw)
#Then display images side by side
numpy.seterr(divide="ignore") #remove warning messages from numpy
figure(figsize=(12,6))
subplot(1,2,1)
imshow(numpy.log(raw), interpolation="nearest", origin="lower")
subplot(1,2,2)
imshow(numpy.log(cor), interpolation="nearest", origin="lower")
ERROR:pyFAI.distortion:The image shape ((1024, 2048)) is not the same as the detector ((1025, 2048)). Adapting shape ...
<matplotlib.image.AxesImage at 0x7f22fb6d48d0>
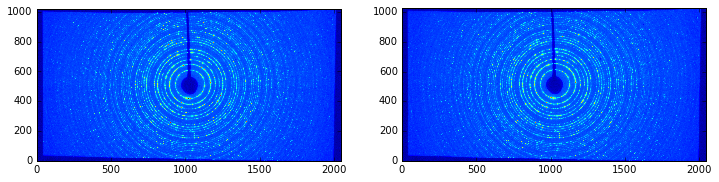
Nota: in this case the image size (1024 lines) does not match the detector’s number of lines (1025) hence pyFAI complains about it. Here, pyFAI patched the image on an empty image of the right size so that the processing can occur.
In this example, the size of the pixels and the shape of the detector are preserved, discarding all pixels falling outside the detector’s grid.
One may want all pixels’ intensity to be preserved in the transformation. By allowing the output array to be large enough to accomodate all pixels, the total intensity can be kept. For this, just enable the “resize” option in the constructor of Distortion:
dis1 = Distortion(hd, resize=True)
print(dis1)
cor = dis1.correct(raw)
print(dis1)
print("After correction, the image has a different shape", cor.shape)
ERROR:pyFAI.distortion:The image shape ((1024, 2048)) is not the same as the detector ((1025, 2048)). Adapting shape ...
Distortion correction lut on device None for detector shape None:
Detector FReLoN Spline= /users/kieffer/workspace-400/pyFAI/doc/source/usage/tutorial/Distortion/halfccd.spline PixelSize= 4.842e-05, 4.684e-05 m
Distortion correction lut on device None for detector shape (1045, 2052):
Detector FReLoN Spline= /users/kieffer/workspace-400/pyFAI/doc/source/usage/tutorial/Distortion/halfccd.spline PixelSize= 4.842e-05, 4.684e-05 m
After correction, the image has a different shape (1045, 2052)
Example of Pixel-detectors:¶
XPad Flat detector¶
There is a striking example in the cover image of this article: http://scripts.iucr.org/cgi-bin/paper?S1600576715004306 where a detector made of multiple modules is eating up some rings. The first example will be about the regeneration of an “eyes friendly” version of this image.
xpad_url = "http://www.silx.org/pub/pyFAI/testimages/LaB6_18.57keV_frame_13.edf"
xpad_file = download(xpad_url)
xpad = pyFAI.detector_factory("Xpad_flat")
print(xpad)
xpad_dis = Distortion(xpad, resize=True)
raw = fabio.open(xpad_file).data
cor = xpad_dis.correct(raw)
print("Shape as input and output:", raw.shape, cor.shape)
#then display images side by side
figure(figsize=(12,10))
subplot(1,2,1)
imshow(numpy.log(raw), interpolation="nearest", origin="lower")
subplot(1,2,2)
imshow(numpy.log(cor), interpolation="nearest", origin="lower")
print("Conservation of the total intensity:", raw.sum(), cor.sum())
Detector Xpad S540 flat PixelSize= 1.300e-04, 1.300e-04 m
Shape as input and output: (960, 560) (1174, 578)
Conservation of the total intensity: 11120798 1.11208e+07

WOS XPad detector¶
This is a new WAXS opened for SAXS pixel detector from ImXPad (available at ESRF-BM02/D2AM CRG beamline). It looks like two of XPad_flat detectors side by side with some modules shifted in order to create a hole to accomodate a flight-tube which gathers the SAXS photons to a second detector further away.
The detector definition for this specific detector has directly been put down using the metrology informations from the manufacturer and saved as a NeXus detector definition file.
wos_det = download("http://www.silx.org/pub/pyFAI/testimages/WOS.h5")
wos_img = download("http://www.silx.org/pub/pyFAI/testimages/WOS.edf")
wos = pyFAI.detector_factory(wos_det)
print(wos)
wos_dis = Distortion(wos, resize=True)
raw = fabio.open(wos_img).data
cor = wos_dis.correct(raw)
print("Shape as input and output:", raw.shape, cor.shape)
#then display images side by side
figure(figsize=(12,10))
subplot(1,2,1)
imshow(numpy.log(raw), interpolation="nearest", origin="lower")
subplot(1,2,2)
imshow(numpy.log(cor), interpolation="nearest", origin="lower")
print("Conservation of the total intensity:", raw.sum(), cor.sum())
NexusDetector detector from NeXus file: WOS.h5 PixelSize= 1.300e-04, 1.300e-04 m
Shape as input and output: (598, 1154) (710, 1302)
Conservation of the total intensity: 444356428 4.44363e+08
/scisoft/users/jupyter/jupy34/lib/python3.4/site-packages/ipykernel/__main__.py:14: RuntimeWarning: invalid value encountered in log
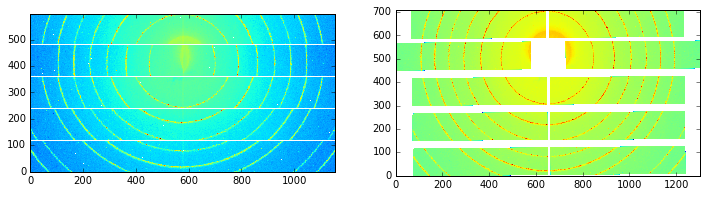
Nota: Do not use this detector definition file to process data from the WOS@D2AM as it has not (yet) been fully validated and may contain some errors in the pixel positioning.
Conclusion¶
PyFAI provides a very comprehensive list of detector definitions, is versatile enough to address most area detectors on the market, and features a powerful regridding engine, both combined together into the distortion correction tool which ensures the conservation of the signal during the transformation (the number of photons counted is preserved during the transformation)
Distortion correction should not be used for pre-processing images prior to azimuthal integration as it re-bins the image, thus induces a broadening of the peaks. The AzimuthalIntegrator object performs all this together with integration, it has hence a better precision.
This tutorial did not answer the question how to calibrate the distortion of a given detector ? which is addressed in another tutorial called detector calibration.