Stitching  ¶
¶
Stitching is the action of agregating together several datasets or several reconstructed volumes. This is a brief overview of stitching methods and how to benefit from it.
z-stitching¶
pre processing z-stitching¶
You can stitch together a z-serie in pre processing by using the PreProcessZStitcher.
It requires as input a set of Nxtomo.
from the command line interface¶
you need first to create a configuration file for stitching.¶
this can be done from:
nabu-stitching-config
you can specify the option level to be droped from the level option and specify an output file. (.conf extension is recommanded).
In the future you should be able to the type of stitching to apply from the stitching_type option.
Once this is done you must edit the file before calling the nabu-stitching application from it.
Some description of the vertical_overlap_area and stitching_height are provided further.
call nabu-stitching on a stitching configuration file.¶
Once you finished editing you stithcing configuration file you can xecute the stitching from:
nabu-stitching [my_stitching_file]
This will trigger the processing.
If this is a pre-processing stitching then this will generate a NXtomo that you will be able to reconstruct using the nabu command line and a nabu reconstruction file.
If this is a post-processing stitching then you will obtain a volume.
from python and notebook¶
to ease processing stitching and get a better view of the behavior there is two notebooks that could be used:
preprocess_stitching: to ease full preprocessing stitching, display some raw data and some basic stiching
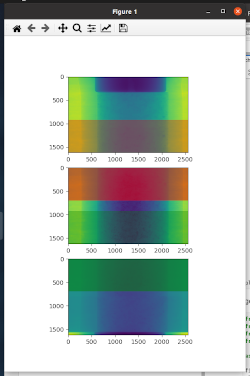
overlap_stitcher: to display and ease definition of overlap areas and result on a single slice. Here is an example on how the overlap areas will be displayed (colored rectangle of alpha==0.5 are the one that can be used for overlap):
note on calling nabu with the stitched NXtomo¶
Pre processing z-stitching will create a NXtomo containing projection already flat fielded. This is why you should turn of flatfield field from the nabu configuration file. as it does not contain any dark or flat it would raise an error otherwise.
note on ordering¶
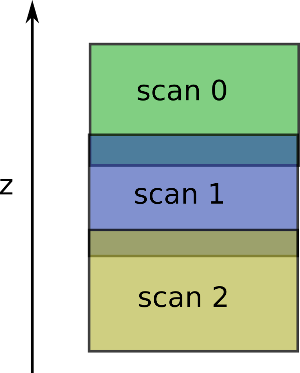
Algorithm expects to have the scan z decreasing ordered (first scan provided should be the upper one).
Only the ZStitcher and the nabu-stiching can invert only scan z ordering automatically.
It expect to have something like:
note on frame flips¶
The NXtomo can be aware of some frame flipping in x or in y. Those flips will be handled by the stitching algorithm. But be aware that stitching algorithm will generate unflipped data. This might be a bit disturbing when comparing stitched data with raw data.
management of rotation angle¶
some acquisition can have inverted rotation angle. To speed up acquition over several z then user might want to avoid to reset to original position motors. This case is hanled by hckecing the order as long as rotation angle are coherent.
overlap areas and z shift.¶
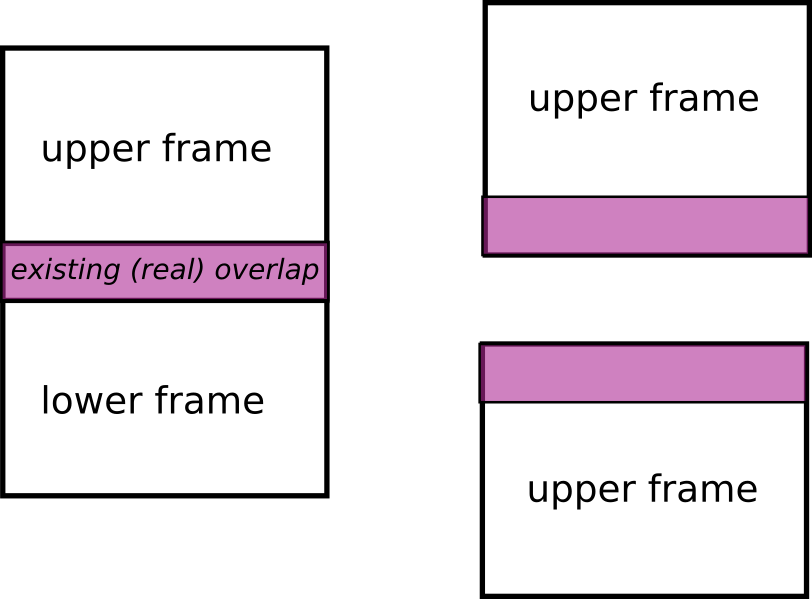
Algorithm expects to have an overlap and a y shift area. Otherwise it will raise an error.
The overlap is the real overlap between two scans. For now it does not handle the case were it does not exists.
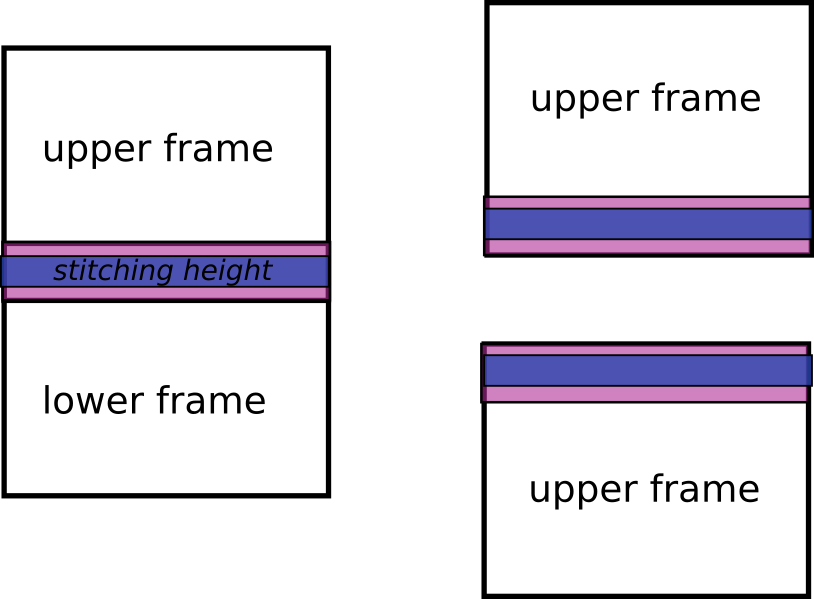
Then on this region user can define a subset of this region (stitching height) to reduce the section to be used for stitching. If this height is not defined then the largest section will be take (stitching height == real overlap).
design¶
All the stitching feature is embed inside nabu stitching module. To split complexity and test behavior we created the following classes:
Stitcher classes (:class:
ZStitcher, :class:PreProcessZStitcher): define the high level API and behavior of the stitcher. Take url as input and volume or NXtomo as output.Overlap class (:class:
ZStichOverlapKernel) class to define overlap between two scans and overlap stratefy classes (:class:OverlapStichingStrategy)Frame composition utils (:class:
ZFrameComposition): ease understanding of the stitching.utils function:
:func:
z_stitch_raw_frames: raw stitching of several frames:func:
z_stitching: high level function to apply stitching according to a user defined configuration
possible optimization to come:¶
speeding up io
write result into several file and create virtual dataset from it
vectorize (some?) calls to
ZStitcher.stitch_frames