Introduction to NumPy¶
- Why do we need NumPy?
- NumPy overview
- The NumPy array
- Creation
- Save and load
- Manipulation
- Indexing
- Copy vs view
- NumPy modules
Why do we need NumPy?¶
Python basic types¶
- Numbers:
10, 10.0, 1.0e+01, (10.0+3j) - Strings:
"Hello world" - Bytes:
b"Hello world" - Lists:
["abc", 3, "x"] - Tuples:
("abc", 3, "x") - Dictionnaries:
{"key1": "abc", "key2": 3, "key3": "x"}
Python basic operators¶
+: Addition-: Substraction/: Division**: Exponentiationabs(x): Absolute value of xx % y: Remainder of x divided by yx // y: Quotient of the x divided by y
Operations on basic Python types¶
# Let a be a list
a = [1, 2, 3]
print(a)
# What is the results of
2 * a[2]
# and
2 * a
# is [2, 4, 6] the expected result?
# You can try other combinations of operations and data types.
Without additional libraries Python is almost useless for scientific computing.
Scientist's Swiss Army knife¶
- Numpy provides support for large, multi-dimensional arrays and matrices.
- Matplotlib provides support for high quality visualizations.
- SciPy provides additional scientific capabilities.
NumPy overview¶
NumPy is the library providing number crunching capabilities to Python, and enhances Python with tools for:
- treatment of multi-dimensional data
- access to optimized linear algebra libraries
- encapsulation of C and Fortran code
import numpy as np
The NumPy array¶
The np.ndarray object is:
- a collection of elements of the same type
- multidimensional with flexible indexing
- handled as any other Python object
- implemented in memory as a true table optimized for performance
It can be interfaced with other languages.
Array creation¶
# Create an array from a list of values.
a = np.array([1, 2, 3, 5, 7, 11, 13, 17])
a
# Create an array from a list of values and dimensions.
b = np.array([[1, 2, 3], [4, 5, 6]])
b
np.array?
Array creation with dedicated methods¶
Documentation: https://docs.scipy.org/doc/numpy/reference/routines.array-creation.html
np.empty((2, 4))
np.zeros((1, 3))
np.ones((2, 2, 3))
np.arange(start=0, stop=10, step=1)
np.linspace(start=0, stop=10, num=11)
np.identity(2)
Array access¶
a = np.array([1, 2, 3, 5, 7, 11, 13, 17])
# Access the first element
print(a[0])
# Access the second element
print(a[1])
b = np.array([[1, 2, 3], [4, 5, 6]])
# Access first-dimension elements (first row here).
print(b[0])
# Access the second element (column) of the first row.
print(b[0, 1])
Exercise¶
Use Python as a simple calculator and try the basic operations on Python lists and NumPy arrays.
a = [1, 2, 3]
b = np.array(a)
# Python list
print("2 * a[2] =", 2 * a[2])
print("2 * a =", 2 * a)
# NumPy array
print("2 * b[2] =", 2 * b[2])
print("2 * b =", 2 * b)
Types of elements¶
- Integers and real numbers with different precision
- Complex numbers
- Chains of characters
- Any Python object
The the element type can be specified using the dtype argument.
np.zeros((1, 3), dtype=int)
np.arange(3, dtype=np.float64)
a = dict({"key1": 0})
b = [1, 2, 3]
c = "element"
np.array([a, b, c], dtype=object)
https://numpy.org/doc/stable/reference/arrays.scalars.html#sized-aliases
- Integers:
int32,int64,uint8... - Real numbers:
float32,float64... - Complex:
complex64,complex128
a = np.arange(10, dtype=np.float32)
print(f"The size of the array is {a.size * a.itemsize} bytes.")
a = np.arange(10, dtype=np.float64)
print(f"The size of the array is {a.size * a.itemsize} bytes.")
Structured/Record arrays¶
They allow access to the data using named fields. Imagine your data being a spreadsheet, the field names would be the column heading.
img = np.zeros((3,), dtype=[("r", np.float32), ("g", np.float64), ("b", np.int32)])
img
img["r"] = 1.0
img
Save and load¶
Documentation: https://docs.scipy.org/doc/numpy/reference/routines.io.html
a = np.arange(start=0, stop=10, step=1, dtype=np.int32)
a
# Save as a binary file (.npy).
np.save("data.npy", a)
np.load("data.npy")
# Save as a text file.
np.savetxt("myarray.txt", a, fmt="%d")
!cat myarray.txt
np.loadtxt("myarray.txt", dtype=np.int32)
Plotting NumPy arrays using Matplotlib¶
Matplotlib is a versatile plotting library that can be used to produce high-quality figures. It provides MATLAB-like functions, such as plot and imshow.
Integration in the notebooks can be enabled using %matplotlib magic.
%matplotlib inline
# %matplotlib widget (for interactive plots, but requires the ipympl package)
# %matplotlib nbagg
from matplotlib import pyplot as plt
x = np.array([0.0, 0.33, 0.66, 0.99, 1.32, 1.65, 1.98, 2.31, 2.64, 2.98, 3.31, 3.64, 3.97, 4.3, 4.63, 4.96, 5.29, 5.62, 5.95, 6.28])
y = np.array([ 1.0, 0.94, 0.79, 0.55, 0.24, -0.08, -0.4, -0.68, -0.88, -0.99, -0.99, -0.88, -0.68, -0.4, -0.08, 0.24, 0.55, 0.79, 0.95, 1.0])
fig = plt.figure()
plt.plot(y)
# plt.plot(x, y)
image = np.random.rand(100, 50)
plt.imshow(image)
plt.colorbar()
Exercise¶
Open the HPLC exercise notebook
Manipulation¶
Array operations¶
Common functions are:
- Linear algebra:
matmulmatrix multiplication,dotproduct,innerproduct,outerproduct - Statistics:
mean,std,median,percentile, ... (https://docs.scipy.org/doc/numpy/reference/routines.statistics.html) - Sums:
sum,cumsum, ... - Math:
cos,sin,log10,interp, ... (https://docs.scipy.org/doc/numpy/reference/routines.math.html) - Indexing, logic functions, sorting
- See: https://docs.scipy.org/doc/numpy/reference/routines.html
a = np.linspace(0.0, 1.0, 100)
print("Mean:", np.mean(a), ", Standard deviation:", np.std(a))
# Standard operations operate element by element.
angles = np.linspace(0, np.pi, 5)
np.cos(angles)
a = np.array([[0.0, 1.0, 2.0], [3.0, 4.0, 5.0], [6.0, 7.0, 8.0]])
b = np.identity(3)
np.matmul(a, b) # Or equivalently a @ b
Array operations along an axis¶
Many NumPy reduction functions take an axis argument.
a = np.array([[0, 1, 2, 3], [4, 5, 6, 7]])
a
np.min(a)
np.min(a, axis=1)
Array methods¶
Some Numpy functions are also available as methods.
a = np.array([[7, 6, 5, 4], [3, 2, 1, 0]])
a
# Returns a value computed from the array
a.min(), a.max(), a.sum()
# An in-place sort operation.
a.sort(axis=1)
a
More on array methods¶
a = np.array([(0, 1), (2, 3)])
a
a.transpose()
np.transpose(a)
b = a.copy()
c = np.copy(a)
d = np.array(a, copy=True)
Be careful when using copy as it is shallow, and it will not copy object elements within arrays. For this, you need to use copy.deepcopy.
a = np.array([1, "m", [2, 3, 4]], dtype=object)
c = np.copy(a)
a[0] = 2
a[2][0] = -1
a, c
Array attributes¶
The dtype attribute identifies the type of the elements of the array.
a = np.array([[3, 2], [8, 12]])
a.dtype
a.dtype.name, a.dtype.str
The shape attribute is a tuple containing the array dimensions.
a = np.array([1, 2, 3, 4])
a.shape
# It can also be set.
a.shape = (2, 2)
a
More array attributes¶
ndim: Number of dimensionssize: Total number of elementsitemsize: Size of a single itemstrides: Bytes to step in each dimensionflags: Contiguity of the data in the buffernbytes: Size in bytes occupied in memorydata: Read/write buffer containing the data
a = np.array(
[[1, 2],
[3, 4]])
a.ndim
Exercise¶
Continue the HPLC exercise notebook - Part II
Indexing¶
Select elements as with any other Python sequence.
- Indexing starts at
0for each array dimension - Indexes can be negative:
x[-1]is the same asx[len(x) - 1]
a = np.array([0, 1, 2, 3])
print("a[0] =", a[0])
print("a[-1] =", a[-1])
a = np.array([(1, 2, 3, 4), (5, 6, 7, 8), (9, 10, 11, 12)])
a
a[2] # Select all the elements of the third row.
a[2, :] # Same as previous, assuming the array has at least two dimensions.
a[1, 2] # Select the element from the second row and third column.
a[0, -1] # Select the last element of the first row.
a[0:2, 0:4:2] # More elaborate indexing using the `start:stop:step` syntax.
More indexing¶
a = np.arange(10.0, 18.0)
a
# The index argument can be a list or an array.
a[[0, 3, 5]]
# The index argument can be a logical array.
mask = a > 13
print("a > 13 =", mask)
a[mask]
Assignment¶
a
a[0:2] = 5 # Assign new values to array elements.
a
Exercise¶
Continue the HPLC exercise notebook - Part III
Exercise¶
- Calculate the element-wise difference between
xandy? - Provide an expression to calculate the difference
x[i+1]-x[i]for all the elements of the 1D array.
x = np.arange(10)
y = np.arange(1, 11)
print("x =", x)
print("y =", y)
# TODO
import exercicesolution
exercicesolution.show("ex3_1")
exercicesolution.show("ex3_2")
Copy vs view¶
You saw previously data copy. But you can work on the same raw data with different views (representations).
- copy: duplicate the data
- view: new array object pointing to the same data

a = np.array([[0, 1], [2, 3]])
b = a.transpose()
a, b


c = a[0]
c

d = a.copy()
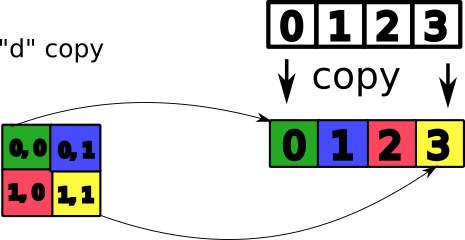
a[0, 0] = 4
print("a:", a)
print("b:", b)
print("c:", c)
print("d:", d)
Exercise¶
Perform a 2x2 binning of an image
Binning with a 1D array
- 1.1: Generate a 1D array with 100 elements in increasing order
- 1.2: Perform a binning such that:
raw data: 1 2 3 4
binned data: 1+2 3+4
Binning with a 2D array 2x2 binning
- 2.1: Generate a 100x100 2D array with elements in increasing order
- 2.2: Perform a binning such that:
| 1 | 2 | 3 | 4 |
|---|---|---|---|
| 5 | 6 | 7 | 8 |
| 9 | 10 | 11 | 12 |
| 13 | 14 | 15 | 16 |
| 1+2+5+6 | 3+4+7+8 |
|---|---|
| 9+10+13+14 | 11+12+15+16 |
- Set all elements of the resulting array that are below 1000 to 0.
# TODO
import exercicesolution
exercicesolution.show("ex4_1")
exercicesolution.show("ex4_2")
exercicesolution.show("ex4_2_alt")
NumPy modules¶
Documentation: https://docs.scipy.org/doc/numpy/reference
Linear algebra: numpy.linalg¶
numpy.linalg.det(x): determinant of xnumpy.linalg.eig(x): eigenvalues and eigenvectors of xnumpy.linalg.inv(x): inverse matrix of xnumpy.linalg.svd(x): singular value decomposition of x
np.linalg.det?
help(np.linalg.det)
Random sampling: numpy.random¶
Simple random data¶
# Random integers in the interval [low:high)
np.random.randint(low=0, high=5, size=10)
# Random floats in the interval [0.0:1.0)
np.random.random(10)
np.random.bytes(10)
Permutations¶
a = np.arange(1, 10)
a
# In-place element permutation
np.random.shuffle(a)
a
# Out-of-place permutation
np.random.permutation(a)
Statistical distributions¶
Normal (Gaussian), Poisson, etc.
data = np.random.normal(loc=1.0, scale=1.0, size=100000)
%matplotlib inline
from matplotlib import pyplot as plt
histo, bin_edges = np.histogram(data, bins=100)
bin_centers = (bin_edges[:-1] + bin_edges[1:]) / 2.
plt.plot(bin_centers, histo)
# Or: plt.hist(data, bins=100)
Fast Fourier Transform: numpy.fft¶
numpy.fft.fft: 1D FFTnumpy.fft.fft2: 2D FFTnumpy.fft.fftn: nD FFT
Polynomials: numpy.polynomial¶
In NumPy, polynomials can be created, manipulated, and even fitted. Numpy provides Polynomial, Chebyshev, Legendre, Laguerre, Hermite and HermiteE series.
Exercise¶
- Write a function
fill_array(height, width)to generate an array of dimensions (height, width) in whichX[row, column] = cos(row) * sin(column) - Time-it for height=1000, width=1000
Bonus: Do the same for X[row, column] = cos(row) + sin(column)
def fill_array(height, width):
a = np
%timeit fill_array(1000, 1000)
# inefficient fill
import exercicesolution
exercicesolution.show("ex5_inefficient_fill")
%timeit exercicesolution.ex5_inefficient_fill(1000, 1000)
# naive fill
exercicesolution.show("ex5_naive_fill")
%timeit exercicesolution.ex5_naive_fill(1000, 1000)
# clever fill
exercicesolution.show("ex5_clever_fill")
%timeit exercicesolution.ex5_clever_fill(1000, 1000)
# practical fill
exercicesolution.show("ex5_practical_fill")
%timeit exercicesolution.ex5_practical_fill(1000, 1000)
# optimized fill
exercicesolution.show("ex5_optimized_fill")
%timeit exercicesolution.ex5_optimized_fill(1000, 1000)
# atleast_2d fill
exercicesolution.show("ex5_atleast_2d_fill")
%timeit exercicesolution.ex5_atleast_2d_fill(1000, 1000)
Speed is a question of algorithm. It is not just a question of language.
| Implementation | Duration (seconds) |
|---|---|
| ex5_inefficient_fill | 5.052937 |
| ex5_naive_fill | 0.886003 |
| ex5_clever_fill | 0.016836 |
| ex5_practical_fill | 0.014959 |
| ex5_optimized_fill | 0.004497 |
| ex5_atleast_2d_fill | 0.005262 |
Done on Intel(R) Xeon(R) CPU E5-1650 @ 3.50GHz
Additional resources¶
- Complete reference material: http://docs.scipy.org/doc/numpy/reference
- NumPy user guide: https://docs.scipy.org/doc/numpy/user
- Many recipes for different purposes: https://scipy-cookbook.readthedocs.io
- Active mailing list where you can ask your questions: numpy-discussion@scipy.org
- Internal data-analysis mailing list: data-analysis@esrf.fr
More exercises for the braves¶
Thanks to Nicolas Rougier: https://github.com/rougier/numpy-100:
- Create a 5x5 matrix with values 1,2,3,4 just below the diagonal.
- Create a 8x8 matrix and fill it with a checkerboard pattern.
- Normalize a 5x5 random matrix.
- Create a 5x5 matrix with row values ranging from 0 to 4.
- Consider a random 10x2 matrix representing cartesian coordinates, convert them to polar coordinates.
- Create random vector of size 10 and replace the maximum value by 0.
- Consider a random vector with shape (100,2) representing coordinates, find point by point distances.
- Generate a generic 2D Gaussian-like array.
- Subtract the mean of each row of a matrix.
- How to I sort an array by the nth column?
- Find the nearest value from a given value in an array.