z-stitching
z-stitching can be done over:
- projections (in this case we will speak of pre-processing stitching)
- reconstructed volumes (in this case we will speak about post-processing stitching)
At the nabu level most of the code is shared for those two use cases. Diversity comes mostly from gathering and interpreting metadata of the scans or of the volumes.
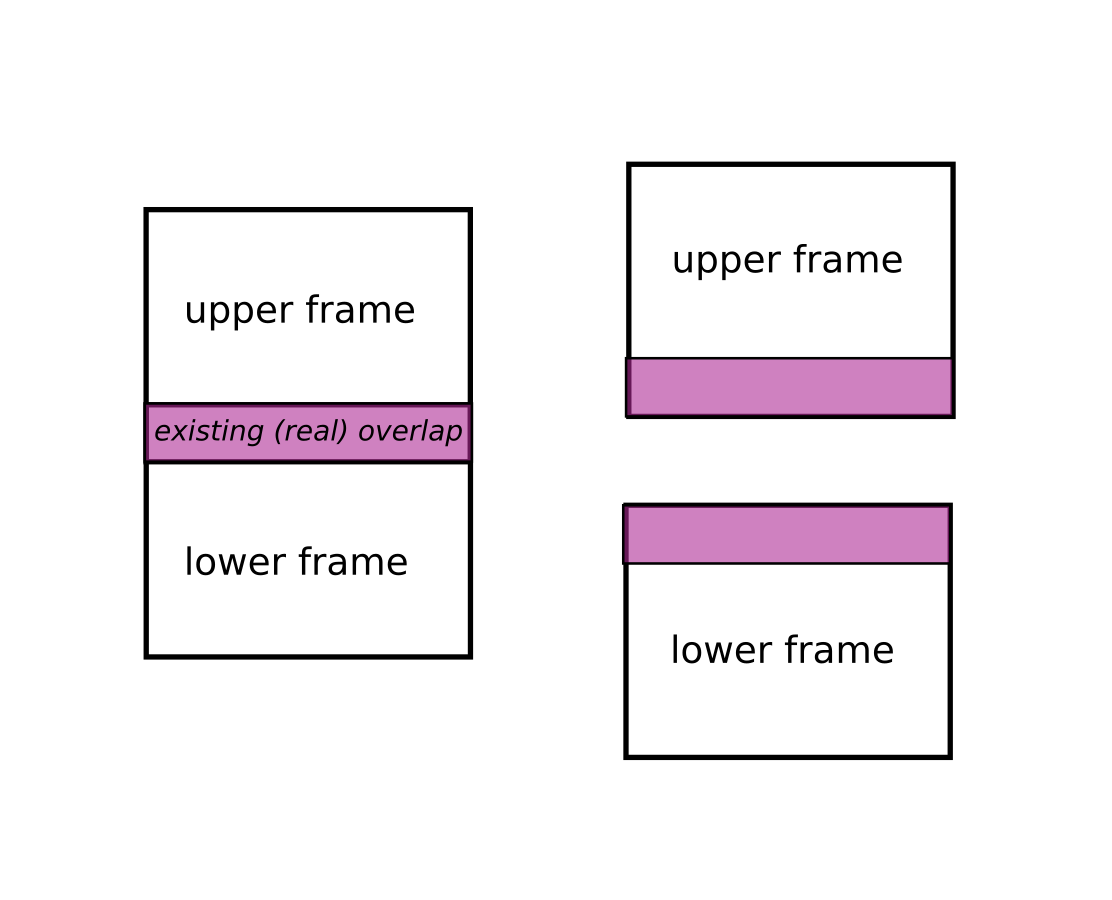
overlapping
To process stitching successfully, frames must have an overlapping area.
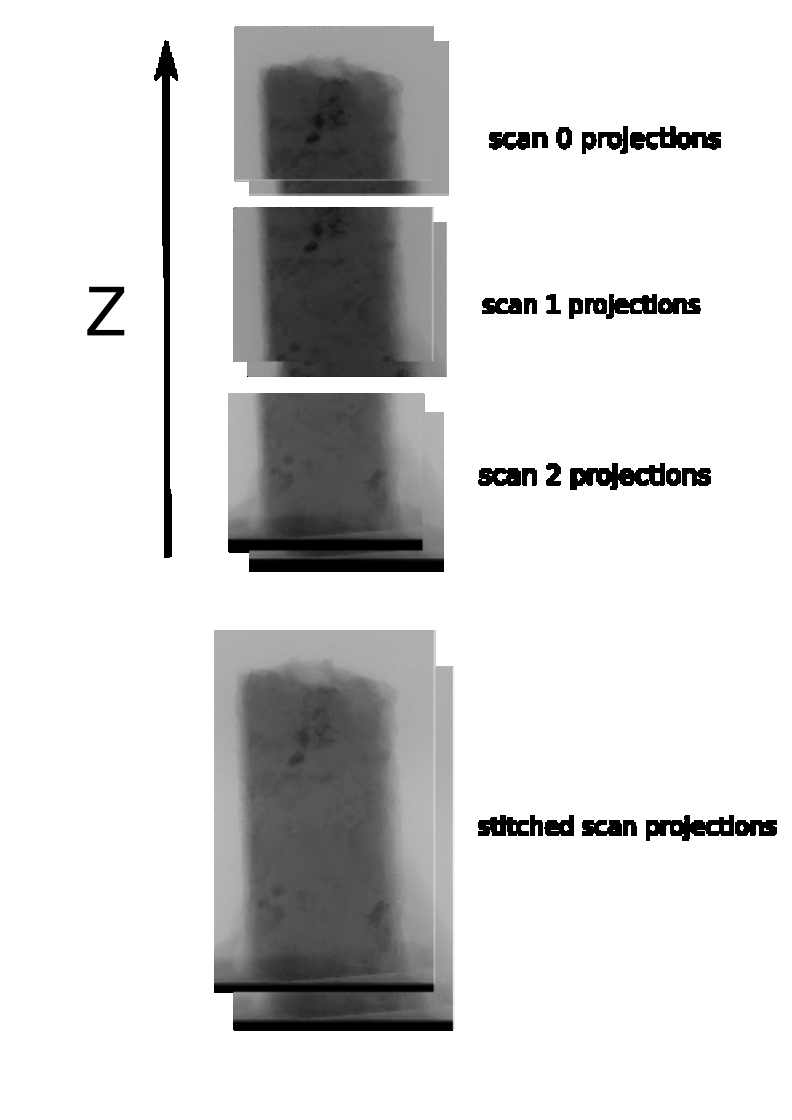
pre processing z-stitching (projections)
You can stitch together a z-series in pre-processing by using the PreProcessZStitcher.
It requires as input a set of NXtomo files.
Real-life example of z-stitching projections
reference used for projection stitching
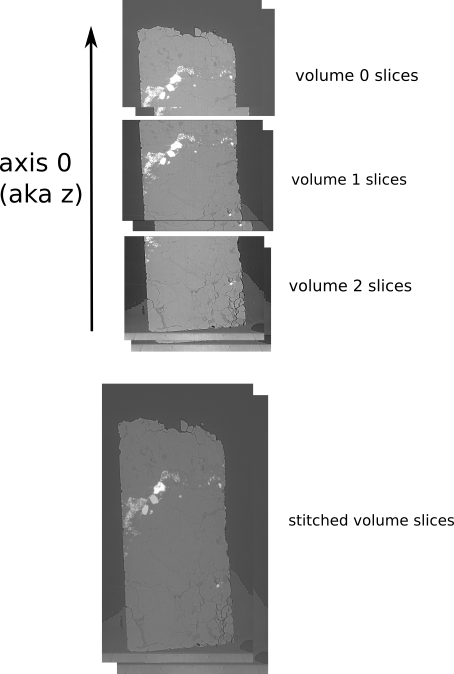
post processing z-stitching (reconstructed volumes)
You can stitch together a z-series in pre-processing by using the PostProcessZStitcher.
It requires as input a set of (reconstructed) volumes.
Real-life example of z-stitching volumes
reference used for volume stitching
stitching using the GUI
A GUI exists to stitch volumes with a graphical user interface. It is embedded with tomwer. See TODO: add link.
stitching from the command line interface
You can apply stitching from the CLI by:
- creating a configuration file
- editing the configuration
- calling
nabu-stitchingwith the configuration file
creating a configuration file
this can be done from:
nabu-stitching-config [-h] [--stitching-type STITCHING_TYPE] [--level LEVEL] [--output OUTPUT]
usage: nabu-stitching-config [-h] [--stitching-type STITCHING_TYPE] [--level LEVEL] [--output OUTPUT]
Initialize a nabu configuration file
optional arguments:
-h, --help show this help message and exit
--stitching-type STITCHING_TYPE
User can provide stitching type to filter some parameters. Must be in ('z-preproc', 'z-postproc').
--level LEVEL Level of options to embed in the configuration file. Can be 'required', 'optional', 'advanced'.
--output OUTPUT output file to store the configuration
editing a configuration file
The different fields of the configuration are explained within the file, but additional precision can be added regarding:
-
input section
slices: You can apply the stitching on a sub-section only (for debugging or distribution for example). This sub-section can be specified using theslicesvalue from the configuration file:
[inputs] slices=slicescan be provided as a slice likeslices=start:stop[:step]or as a list of values likeslices=startorslices=1,20,55. Providing them as slices (with step equal to 1) is recommended because it allows contiguous data to be loaded faster.
calling nabu-stitching with the configuration file
Once you finished editing the stitching configuration file you can execute the stitching from:
nabu-stitching [my_stitching_file]
This will trigger the processing.
If this is a pre-processing stitching then this will generate a NXtomo that you will be able to reconstruct using the nabu command line and a nabu reconstruction file.
If this is a post-processing stitching then you will obtain a volume.
from python and notebook
To ease stitching processing and get a better view of the behavior there are two notebooks that can be used:
- preprocess_stitching: to run full preprocessing stitching, display some raw data and some basic stitching.
- postprocess_stitching: to run full post-processing stitching, display some raw data and some basic stitching.
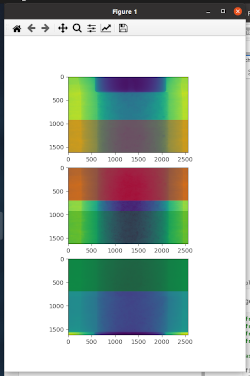
- overlap_stitcher: to display and define overlap areas and results on a single slice. Here is an example of how the overlap areas will be displayed (colored rectangles with alpha == 0.5 indicate regions that can be used for overlap):
note on calling nabu with the stitched NXtomo
Pre-processing z-stitching will create an NXtomo containing projections already flat-fielded. This is why you should turn off the flatfield field in the nabu configuration file. As it does not contain any dark or flat fields it would raise an error otherwise.
note on ordering
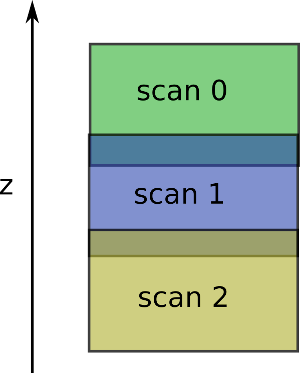
The algorithm expects the scan z positions to be ordered descendingly (the first scan provided should be the upper one).
Only the ZStitcher and the nabu-stitching CLI can automatically invert the scan z ordering.
It expect to have something like:
note on frame flips
The NXtomo can be aware of some frame flipping in x or in y. Those flips will be handled by the stitching algorithm. But be aware that stitching algorithm will generate unflipped data. This might be a bit disturbing when comparing stitched data with raw data.
note on management of rotation angle
Some acquisitions can have inverted rotation angles. To speed up acquisition over several z positions a user might want to avoid resetting the motors to their original position. This case is handled by checking the order as long as rotation angles are coherent.
real overlaps vs stitching overlap.
Between two overlapping scans or volumes there exists a "real overlap", which can be obtained from the real positions of the two objects plus their shapes.
Positions are deduced from the metadata of the object. They can also be redefined by either axis_X_pos_px (one per object) or by axis_X_pos_mm plus the pixel size or voxel size.

Then the algorithm will do the stitching using an overlap size ( <= real overlap). Of course this height must be smaller than or equal to the real overlap otherwise it cannot proceed. This size can be defined by the overlap_size key from the axis_0_params parameter.
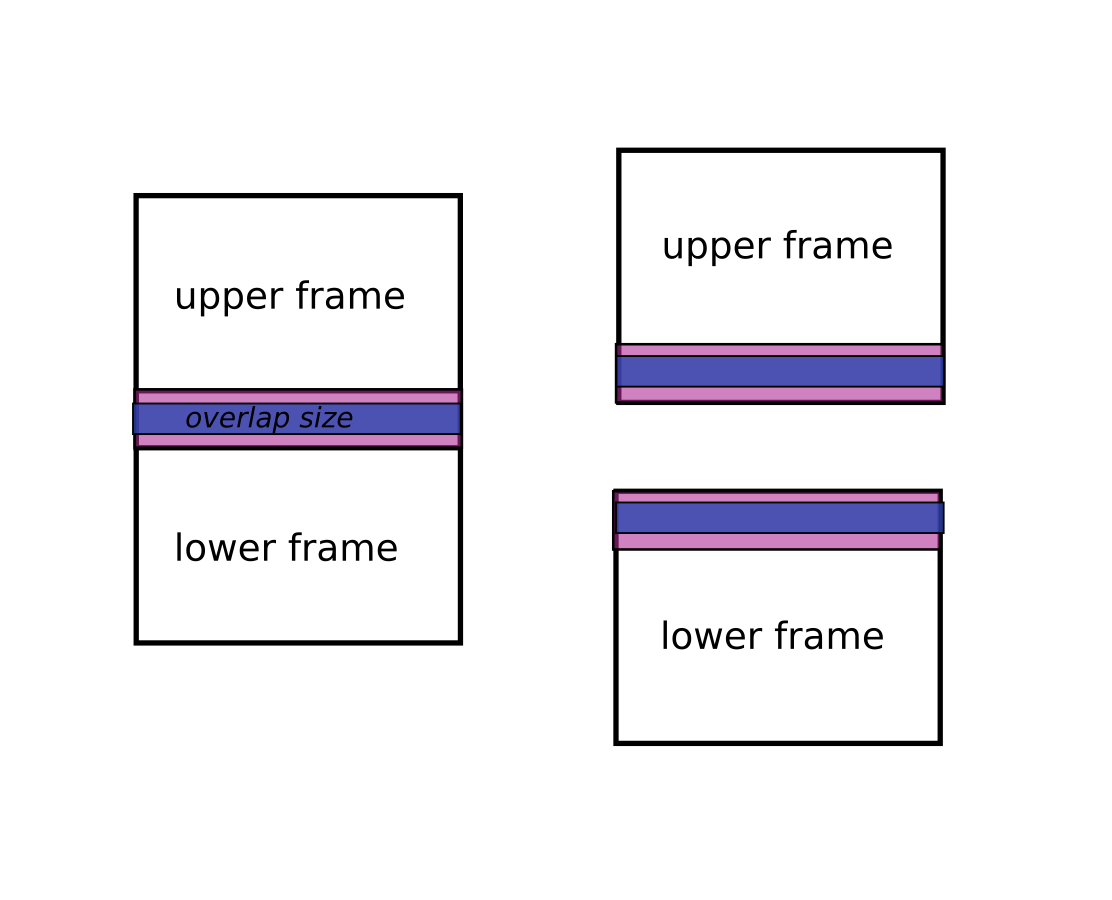
In this example we request the z-stitching overlap to be 40 px high:
axis_0_params = overlap_size=40
warning / bottlenecks
Pre-processing stitching works for the NXtomo format. If your input is an EDF please convert it to NXtomo first. A tutorial can be found here
Post-processing stitching is meant to work with any file format handled by nabu. Nevertheless it primarily targets HDF5. For other file formats it is expected to be unoptimized. For now, for example, it will duplicate all slice files to perform the concatenation.