Orientation management#
Images have 4 corners and most acquisition software offer the ability to flip the image horizontally or vertically leading to 4 possible orientations.
The EXIF specification indicates orientation of each image. The first 4 orientation corresponds to vertical or horzontal flips which change the position of the origin:
Top left when seen from the back of the camera
Top right when seen from the back of the camera , it becomes top-left when looking from the sample.
Bottom right when seen from the back of the camera , it becomes bottom-left when looking from the sample.
Bottom left when seen from the back of the camera
transposed image: unsupported
rotated image: unsupported
transposed image: unsupported
rotated image: unsupported
PyFAI does not support the images which are rotated (90° or 270°) or transposed images. The default orientation is 3 with the origin at the Bottom-Right (Bottom-Left when seen from the sample).
This tutorial demonstrates the image flipping and how it can be accounted when performing azimuthal integration.
1. Generate images with the 4 orientations:#
%matplotlib inline
import time
import numpy
from matplotlib.pyplot import subplots
import pyFAI
from pyFAI.gui import jupyter
from pyFAI.test.utilstest import UtilsTest
t0 = time.perf_counter()
print(f"pyFAI version: {pyFAI.version}")
unit = "q_nm^-1"
pyFAI version: 2025.12.0
import fabio
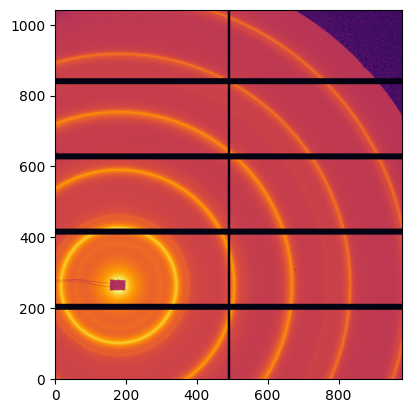
img = fabio.open(UtilsTest.getimage("Pilatus1M.edf")).data
jupyter.display(img);
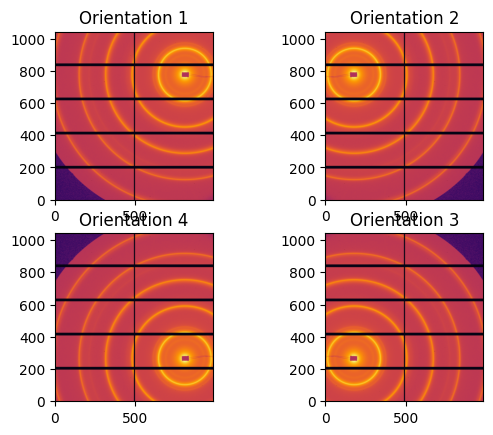
fig, ax = subplots(2, 2)
images = {3:img}
images[2] = numpy.flipud(img)
images[4] = numpy.fliplr(img)
images[1] = numpy.fliplr(images[2])
jupyter.display(images[1], ax=ax[0,0]).set_title("Orientation 1")
jupyter.display(images[2], ax=ax[0,1]).set_title("Orientation 2")
jupyter.display(images[3], ax=ax[1,1]).set_title("Orientation 3")
jupyter.display(images[4], ax=ax[1,0]).set_title("Orientation 4");
Initalize azimuthal integrator with different orientation:#
ai = pyFAI.load(UtilsTest.getimage("Pilatus1M.poni"))
ais = {}
for o in range(1,5):
aix = ai.__copy__()
aix.detector = pyFAI.detector_factory("Pilatus1M", {"orientation": o})
ais[o] = aix
ais
{1: Detector Pilatus 1M PixelSize= 172µm, 172µm TopLeft (1)
Wavelength= 1.000000 Å
SampleDetDist= 1.583231e+00 m PONI= 3.341702e-02, 4.122778e-02 m rot1=0.006487 rot2=0.007558 rot3=0.000000 rad
DirectBeamDist= 1583.310 mm Center: x=179.981, y=263.859 pix Tilt= 0.571° tiltPlanRotation= 130.640° λ= 1.000Å,
2: Detector Pilatus 1M PixelSize= 172µm, 172µm TopRight (2)
Wavelength= 1.000000 Å
SampleDetDist= 1.583231e+00 m PONI= 3.341702e-02, 4.122778e-02 m rot1=0.006487 rot2=0.007558 rot3=0.000000 rad
DirectBeamDist= 1583.310 mm Center: x=179.981, y=263.859 pix Tilt= 0.571° tiltPlanRotation= 130.640° λ= 1.000Å,
3: Detector Pilatus 1M PixelSize= 172µm, 172µm BottomRight (3)
Wavelength= 1.000000 Å
SampleDetDist= 1.583231e+00 m PONI= 3.341702e-02, 4.122778e-02 m rot1=0.006487 rot2=0.007558 rot3=0.000000 rad
DirectBeamDist= 1583.310 mm Center: x=179.981, y=263.859 pix Tilt= 0.571° tiltPlanRotation= 130.640° λ= 1.000Å,
4: Detector Pilatus 1M PixelSize= 172µm, 172µm BottomLeft (4)
Wavelength= 1.000000 Å
SampleDetDist= 1.583231e+00 m PONI= 3.341702e-02, 4.122778e-02 m rot1=0.006487 rot2=0.007558 rot3=0.000000 rad
DirectBeamDist= 1583.310 mm Center: x=179.981, y=263.859 pix Tilt= 0.571° tiltPlanRotation= 130.640° λ= 1.000Å}
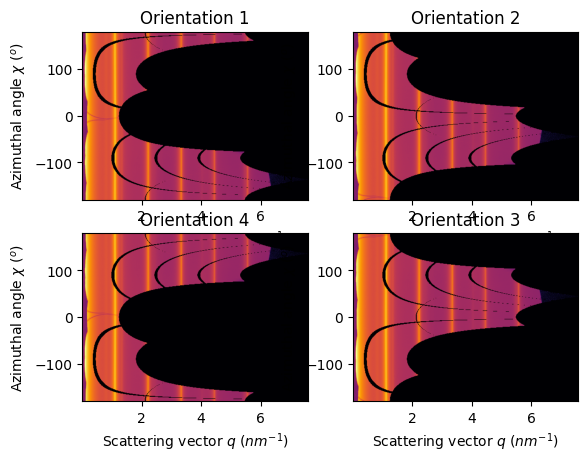
Perform the azimuthal integration#
# With full pixel splitting:
method = ("full", "histogram", "cython")
fig, ax = subplots(2, 2)
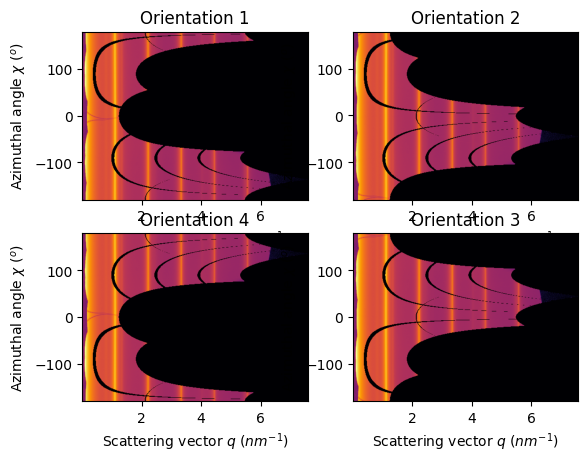
jupyter.plot2d(ais[1].integrate2d(images[1],500, 360, method=method, unit=unit), ax=ax[0,0]).set_title("Orientation 1")
jupyter.plot2d(ais[2].integrate2d(images[2],500, 360, method=method, unit=unit), ax=ax[0,1]).set_title("Orientation 2")
jupyter.plot2d(ais[3].integrate2d(images[3],500, 360, method=method, unit=unit), ax=ax[1,1]).set_title("Orientation 3")
jupyter.plot2d(ais[4].integrate2d(images[4],500, 360, method=method, unit=unit), ax=ax[1,0]).set_title("Orientation 4");
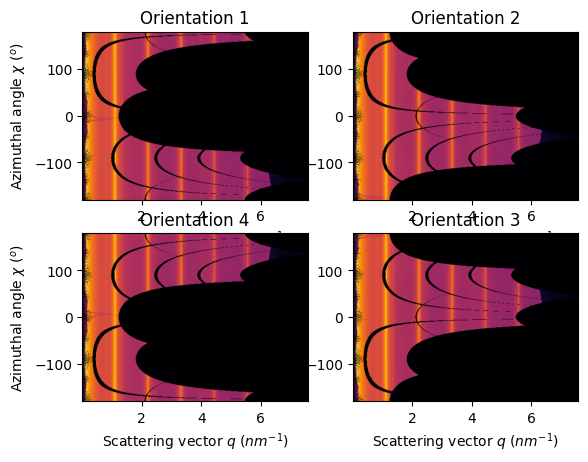
# Without pixel splitting:
method = ("no", "histogram", "cython")
fig, ax = subplots(2, 2)
jupyter.plot2d(ais[1].integrate2d(images[1],500, 360, method=method, unit=unit), ax=ax[0,0]).set_title("Orientation 1")
jupyter.plot2d(ais[2].integrate2d(images[2],500, 360, method=method, unit=unit), ax=ax[0,1]).set_title("Orientation 2")
jupyter.plot2d(ais[3].integrate2d(images[3],500, 360, method=method, unit=unit), ax=ax[1,1]).set_title("Orientation 3")
jupyter.plot2d(ais[4].integrate2d(images[4],500, 360, method=method, unit=unit), ax=ax[1,0]).set_title("Orientation 4");
# Without bounding-box pixel splitting:
method = ("bbox", "histogram", "cython")
fig, ax = subplots(2, 2)
jupyter.plot2d(ais[1].integrate2d(images[1], 500, 360, method=method, unit=unit), ax=ax[0,0]).set_title("Orientation 1")
jupyter.plot2d(ais[2].integrate2d(images[2], 500, 360, method=method, unit=unit), ax=ax[0,1]).set_title("Orientation 2")
jupyter.plot2d(ais[3].integrate2d(images[3], 500, 360, method=method, unit=unit), ax=ax[1,1]).set_title("Orientation 3")
jupyter.plot2d(ais[4].integrate2d(images[4], 500, 360, method=method, unit=unit), ax=ax[1,0]).set_title("Orientation 4");
print(f"Total run-time: {time.perf_counter()-t0:.3f}s")
Total run-time: 6.723s
Conclusion#
The PONI is valid from one geometry to another and the 1d azimuthal integration is the same. But the azimuthal angle does vary thus the 2D integration is mirrored for orientation 2 and 4 and offset by 180° for orientation 1.