Pixel splitting#
This notebook demonstrates the layout of pixel in polar coordinates on a small detector (5x5 pixels) to demonstrate pixel splitting and pixel reorganisation.
In a second part, it displays the effect of the splitting scheme on 2D integration.
# %matplotlib widget
#For documentation purpose, `inline` is used to enforce the storage of the image in the notebook
%matplotlib inline
import time
import numpy
from matplotlib.pyplot import subplots
from matplotlib.patches import Polygon
from matplotlib.collections import PatchCollection
start_time = time.perf_counter()
import fabio
import pyFAI.test.utilstest
from pyFAI.gui import jupyter
import pyFAI, pyFAI.units
from pyFAI.detectors import Detector
from pyFAI.integrator.azimuthal import AzimuthalIntegrator
from pyFAI.ext import splitPixel
print(f"Using pyFAI version {pyFAI.version}")
Using pyFAI version 2025.12.1-dev0
# Define a dummy 5x5 detector with 1mm pixel size
det = Detector(1e-3, 1e-3, max_shape=(5,5))
print(det)
Detector Detector PixelSize= 1mm, 1mm BottomRight (3)
def area4(a0, a1, b0, b1,c0,c1,d0,d1):
"""
Calculate the area of the ABCD polygon with 4 with corners:
A(a0,a1)
B(b0,b1)
C(c0,c1)
D(d0,d1)
:return: area, i.e. 1/2 * (AC ^ BD)
"""
return 0.5 * (((c0 - a0) * (d1 - b1)) - ((c1 - a1) * (d0 - b0)))
# Example of code for spotting pixel on the azimuthal discontinuity: its area has not the same sign!
chiDiscAtPi = 1
pi = numpy.pi
two_pi = 2*numpy.pi
ai = AzimuthalIntegrator(1, 2.2e-3, 2.8e-3, rot3=-0.3, detector=det)
if chiDiscAtPi:
ai.setChiDiscAtPi()
else:
ai.setChiDiscAtZero()
pos = ai.array_from_unit(typ="corner", unit="r_mm", scale=True)
a = []
s = 0
ss = 0
disc = {}
for i0 in range(pos.shape[0]):
for i1 in range(pos.shape[1]):
p = pos[i0, i1].copy()
area = area4(*p.ravel())
area2 = None
if area>=0:
az = p[:, 1].copy()
if chiDiscAtPi:
m = numpy.where(az<0)
else:
m = numpy.where(az<pi)
az[m] = two_pi + az[m]
c1 = az.mean()
if not chiDiscAtPi and c1>two_pi:
az -= two_pi
elif chiDiscAtPi and c1>pi:
az -= two_pi
disc[(i0, i1)] = list(zip(p.copy(),az))
p[:, 1 ] = az
area2 = area4(*p.ravel())
print(f"({i0} {i1}) A = {area:.3f}" +(f" -> {area2:.3f}" if area2 else ""))
print("Discontinuities")
for k,v in disc.items():
print(k)
for l in v:
print(f" {l[0]} -> {l[1]:.7f}")
(0 0) A = -0.343
(0 1) A = -0.453
(0 2) A = -0.579
(0 3) A = -0.533
(0 4) A = -0.405
(1 0) A = -0.414
(1 1) A = -0.647
(1 2) A = -1.133
(1 3) A = -0.877
(1 4) A = -0.533
(2 0) A = 3.026 -> -0.432
(2 1) A = 4.995 -> -0.738
(2 2) A = 1.791 -> -0.874
(2 3) A = -1.133
(2 4) A = -0.579
(3 0) A = 3.896 -> -0.373
(3 1) A = -0.519
(3 2) A = -0.738
(3 3) A = -0.647
(3 4) A = -0.453
(4 0) A = -0.303
(4 1) A = -0.373
(4 2) A = -0.432
(4 3) A = -0.414
(4 4) A = -0.343
Discontinuities
(2, 0)
[ 2.807134 -2.7702851] -> -2.7702851
[ 2.912044 -3.1198924] -> -3.1198924
[1.9697715 3.0233684] -> -3.2598171
[ 1.811077 -2.7309353] -> -2.7309353
(2, 1)
[ 1.811077 -2.7309353] -> -2.7309353
[1.9697715 3.0233684] -> -3.2598171
[1.1313709 2.6561944] -> -3.6269910
[ 0.82462114 -2.596614 ] -> -2.5966139
(2, 2)
[ 0.82462114 -2.596614 ] -> -2.5966139
[1.1313709 2.6561944] -> -3.6269910
[0.82462114 1.6258177 ] -> -4.6573677
[ 0.28284273 -0.48539817] -> -0.4853983
(3, 0)
[ 2.912044 -3.1198924] -> 3.1632931
[3.3286633 2.8702552] -> 2.8702552
[2.5455844 2.6561944] -> 2.6561944
[1.9697715 3.0233684] -> 3.0233684
The recenteriing of pixels is obtained from pyFAI.ext.splitPixel.recenter which is implemented in Cython to be more efficient.
Beware of the signature changed with pyFAI-2025.12
def display_geometry(pos):
fig, ax = subplots()
patches = []
for i0 in range(pos.shape[0]):
for i1 in range(pos.shape[1]):
p = pos[i0, i1].astype("float64")
splitPixel.recenter(p, chiDiscAtPi=chiDiscAtPi)
p = numpy.concatenate((p, [p[0]]))
ax.plot(p[:,0], p[:,1], "--")
patches.append(Polygon(p))
p = PatchCollection(patches, alpha=0.4)
colors = numpy.linspace(0, 100, len(patches))
p.set_array(colors)
ax.add_collection(p)
if chiDiscAtPi:
ax.plot([0,4], [-pi, -pi])
else:
ax.plot([0,4], [two_pi, two_pi])
ax.plot([0,4], [pi, pi])
ax.plot([0,4], [0, 0])
ax.set_xlabel(unit.label)
ax.set_ylabel("Azimuthal angle (rad)")
return ax
chiDiscAtPi = 0
unit = pyFAI.units.to_unit("r_mm")
ai = AzimuthalIntegrator(1, 2.2e-3, 2.8e-3, rot3=0.5, detector=det)
if chiDiscAtPi:
ai.setChiDiscAtPi()
low = -pi
high = pi
else:
ai.setChiDiscAtZero()
low = 0
high = two_pi
pos = ai.array_from_unit(typ="corner", unit=unit, scale=True)
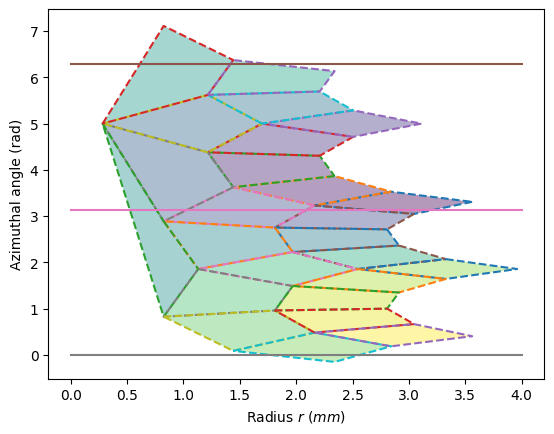
ax = display_geometry(pos)
over = 0
under = 0
for i0 in range(pos.shape[0]):
for i1 in range(pos.shape[1]):
p = pos[i0, i1].copy()
area = area4(*p.ravel())
area2 = None
if area>=0:
az = p[:, 1]
if chiDiscAtPi:
m = numpy.where(az<0)
else:
m = numpy.where(az<pi)
az[m] = two_pi + az[m]
c1 = az.mean()
if not chiDiscAtPi and c1>two_pi:
az -= two_pi
elif chiDiscAtPi and c1>pi:
az -= two_pi
over += (az>high).sum()
under += (az<low).sum()
print(f"Card(χ<{low/pi:.0f}π) = {under} \t Card(χ>{high/pi:.0f}π) = {over}")
Card(χ<0π) = 1 Card(χ>2π) = 3
chiDiscAtPi = 1
pi = numpy.pi
two_pi = 2*numpy.pi
ai = AzimuthalIntegrator(1, 2.2e-3, 2.8e-3, rot3=-0.4, detector=det)
if chiDiscAtPi:
ai.setChiDiscAtPi()
low = -pi
high = pi
else:
ai.setChiDiscAtZero()
low = 0
high = two_pi
pos = ai.array_from_unit(typ="corner", unit="r_mm", scale=True)
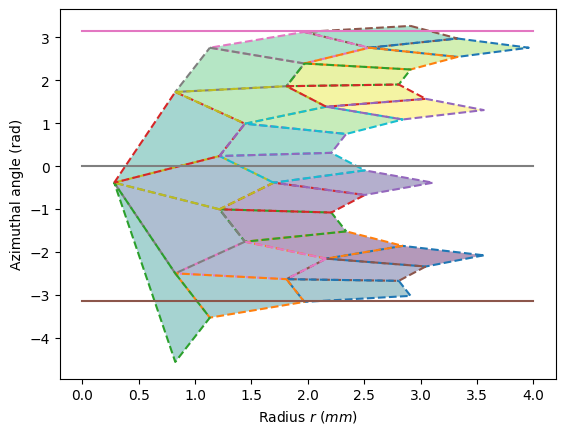
_ = display_geometry(pos)
over = 0
under = 0
for i0 in range(pos.shape[0]):
for i1 in range(pos.shape[1]):
p = pos[i0, i1].copy()
area = area4(*p.ravel())
area2 = None
if area>=0:
az = p[:, 1]
if chiDiscAtPi:
m = numpy.where(az<0)
else:
m = numpy.where(az<pi)
az[m] = two_pi + az[m]
c1 = az.mean()
print(c1)
if c1>high:
az -= two_pi
over += (az>high).sum()
under += (az<low).sum()
print(f"Card(χ<{low/pi:.0f}π) = {under} \t Card(χ>{high/pi:.0f}π) = {over}")
3.412953
3.329596
3.5415926
3.0282776
Card(χ<-1π) = 5 Card(χ>1π) = 1
Effect of pixel splitting on 2D integration#
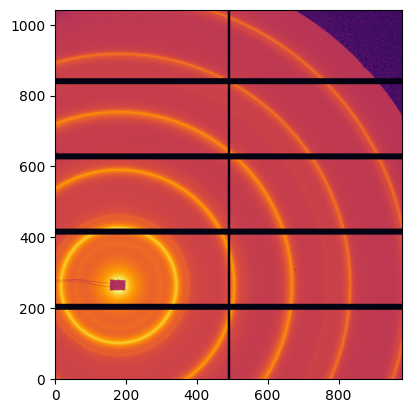
img = pyFAI.test.utilstest.UtilsTest.getimage("Pilatus1M.edf")
fimg = fabio.open(img)
_ = jupyter.display(fimg.data)
# Focus on the beam stop holder ...
poni = pyFAI.test.utilstest.UtilsTest.getimage("Pilatus1M.poni")
ai = pyFAI.load(poni)
print(ai)
ai.setChiDiscAtZero()
Detector Pilatus 1M PixelSize= 172µm, 172µm BottomRight (3)
Wavelength= 1.000000 Å
SampleDetDist= 1.583231e+00 m PONI= 3.341702e-02, 4.122778e-02 m rot1=0.006487 rot2=0.007558 rot3=0.000000 rad
DirectBeamDist= 1583.310 mm Center: x=179.981, y=263.859 pix Tilt= 0.571° tiltPlanRotation= 130.640° λ= 1.000Å
kwargs = {"data":fimg.data,
"npt_rad":500,
"npt_azim":500,
"unit":"r_mm",
"dummy":-2,
"delta_dummy":2,
"azimuth_range":(150,200),
"radial_range":(0,50),
}
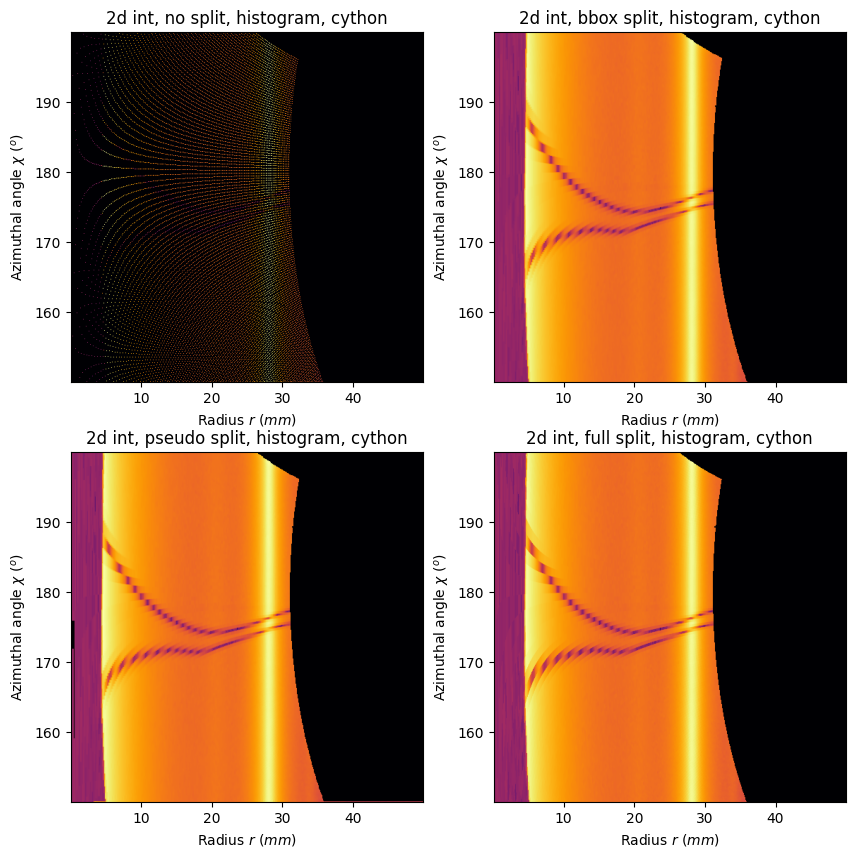
resn = ai.integrate2d_ng(method=("no", "histogram", "cython"), **kwargs)
resb = ai.integrate2d_ng(method=("bbox", "histogram", "cython"), **kwargs)
resp = ai.integrate2d_ng(method=("pseudo", "histogram", "cython"), **kwargs)
resf = ai.integrate2d_ng(method=("full", "histogram", "cython"), **kwargs)
fig,ax = subplots(2,2, figsize=(10,10))
jupyter.plot2d(resn, ax=ax[0,0])
ax[0,0].set_title(resn.compute_engine.split("(")[1].strip(")"))
jupyter.plot2d(resb, ax=ax[0,1])
ax[0,1].set_title(resb.compute_engine.split("(")[1].strip(")"))
jupyter.plot2d(resp, ax=ax[1,0])
ax[1,0].set_title(resp.compute_engine.split("(")[1].strip(")"))
jupyter.plot2d(resf, ax=ax[1,1])
ax[1,1].set_title(resf.compute_engine.split("(")[1].strip(")"))
pass
# Compared performances for 2D integration ...
print("Without pixel splitting")
%timeit ai.integrate2d_ng(method=("no", "histogram", "cython"), **kwargs)
print("Bounding box pixel splitting")
%timeit ai.integrate2d_ng(method=("bbox", "histogram", "cython"), **kwargs)
print("Scalledd Bounding box pixel splitting")
%timeit ai.integrate2d_ng(method=("pseudo", "histogram", "cython"), **kwargs)
print("Full pixel splitting")
%timeit ai.integrate2d_ng(method=("full", "histogram", "cython"), **kwargs)
Without pixel splitting
8.87 ms ± 78.7 μs per loop (mean ± std. dev. of 7 runs, 100 loops each)
Bounding box pixel splitting
16.1 ms ± 16.4 μs per loop (mean ± std. dev. of 7 runs, 100 loops each)
Scalledd Bounding box pixel splitting
23.6 ms ± 35.4 μs per loop (mean ± std. dev. of 7 runs, 10 loops each)
Full pixel splitting
105 ms ± 272 μs per loop (mean ± std. dev. of 7 runs, 10 loops each)
Conclusion#
This tutorial presents how pixels are located in polar space and explains why pixels on the azimuthal discontinuity requires special care. It also presents a comparison between the 4 pixel splitting schemes available in pyFAI: without splitting (no), along the bounding box (bbox), a scaled down bounding box (pseudo) and finally the splitting along the edges of each pixel (full). The corresponding runtimes are also provided.
print(f"runtime: {time.perf_counter()-start_time:.3f}s")
runtime: 33.833s